Acetyl Lysine Antibody, Agarose -2mg
|
catalog#
Product Description
|
ICP0388-2MG
The acetylated lysine antibody is immobilized to beaded agarose via amide linkages. The product could be utilized as an affinity matrix for rapid isolation and purification of proteins and peptides with acetyl lysine residues.
|
|
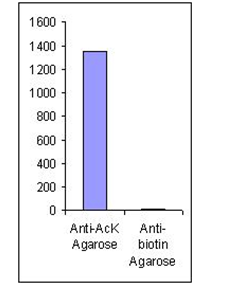 The acetylated peptide profile from acetyl-lysine-specific affinity chromatography (matrix = ICP0388). Approximately 5 mg of trypsinated crude protein from TSA treated MMRU cells were loaded. The acetylated peptide profile from acetyl-lysine-specific affinity chromatography (matrix = ICP0388). Approximately 5 mg of trypsinated crude protein from TSA treated MMRU cells were loaded.
The maximum binding of acetylated BSA with ICP0388-2MG.
-50 µl of ICP0388-2MG were incubated with 1 mg of acetylate BSA in a 1 ml tube for 60 min.
- After washing with PBST 4 times, the bound acetylated BSA was eluted with 1 ml of 0.5 M HCl
-5 µl / lane of the eluted acetylated BSA was resolved by SDS-PAGE and blot wih monoclonal anti-acetylated lysine (ICP0390)
|
| Formulation |
0.5 mL beaded agarose suspended in 1 mL slurry
|
| Antibody Immobilized |
4 mg/ml antibody is covalently linked through amide bonds with NHS activated-SMCC then linked to thiolated agarose beads via thiol ether bonds.
|
| Specificity |
The antibody selectively captures peptides and proteins wtih acetylated lysine residues (N-epsilon). There are no cross reactions wtih methylated proteins or mono- and dimethylated prtoeins.
|
|
Binding
Capacity
|
Approximately 0.2 mg of acetylated histone/ml.
|
| Applications |
Rapid isolation and purification of peptides or proteins with acetylated lysine residues from cell lysate or protease-digested mixtures.
|
| Scientific Description |
Protein acetylation is a form of post-translational modification known to regulate many diverse biological processes. Detection, isolation and identification of acetylated proteins/peptides is essential in proteomic studies. Affinity chromatography is one of the most efficient and rapid methods to enrich and purify the acetylated species for further MS/MS identification.
|
| Storage & Stability |
Product is stable for 30 days at room temperature. For extended storage, store product at -20°C. Do not aliquot and shake thoroughly before use. Expiration date is one year from date of shipping if properly stored.
|
| Product Specific References |
- 1. Science. 2009. 325 (5942): 834-840. doi:10.1126/science.1175371.
- 2. Cell. 2010. 140 (2): 257-267. doi:10.1016/j.cell.2009.12.031.
- 3. Mol. Cell. 2010. 39 (2): 247-258. doi:10.1016/j.molcel.2010.07.006.
- 4. Proteomics. 2010. 10 (5): 1029-1039. doi:10.1002/pmic.200900602.
- 5. Plant Physiol. 2011. 155 (4): 1779-1790. doi:10.1104/pp.110.171595.
- 6. Euro. J. Cell Biol. 2011. 90 (1-2): 128-135. doi:10.1016/j.ejcb.2010.09.004.
- 7. Mol. Cell. Proteomics. 2012. 11 (11): 1510-1522. doi:10.1074/mcp.M112.017251.
- 8. Proc. Natl. Acad. Sci. U.S.A. 2012. 109 (28): 11133-11138. doi:10.1073/pnas.1208669109.
- 9. J. Proteome Res. 2012. 11 (3): 1633-1643. doi:10.1021/pr2008384.
- 10. J. Lipid Res. 2012. 53 (9): 1864-1876. doi:10.1194/jlr.M026567.
- 11. Cell. 2012. 149 (1): 214-231. doi:10.1016/j.cell.2012.02.013.
- 12. Cell.2012. 150 (3): 620-632.doi:10.1016/j.cell.2012.06.027.
- 13. Exp. Hematol. 2012. 40 (4): 342-355. doi:10.1016/j.exphem.2011.12.005.
- 14. PLoS ONE. 2012. 7 (12): e50545. doi:10.1371/journal.pone.0050545.
- 15. PLoS Genet. 2012. 8 (9): e1002948. doi:10.1371/journal.pgen.1002948.
- 16. Euro. J. Cell Biol. 2012. 91 (11-12): 950-960. doi:10.1016/j.ejcb.2012.07.001.
- 17. J. Proteomics.2013. 79: 60-71. doi:10.1016/j.jprot.2012.12.001.
- 18. Biochem. Biophys. Res. Commun. 2013. 435 (3): 403-407. doi:10.1016/j.bbrc.2013.04.101.
- 19. J. Biol. Chem. 2013. 288 (36): 26209-26219. doi:10.1074/jbc.M113.483396.
- 20. J. Biol. Chem. 2013. 288 (40): 29036-29045. doi:10.1074/jbc.M113.486753.
- 21. Mol. Microbiol. 2013. 89 (4): 660-675. doi:10.1111/mmi.12303.
- 22. Proc. Natl. Acad. Sci. U.S.A. 2013. 110 (9): 3339-3344. doi:10.1073/pnas.1217632110.
- 23. Mol. Cell. 2013. 51 (2): 265-272. doi:10.1016/j.molcel.2013.06.003.
- 24. Mol. Cell. Biol. 2013. 33 (8): 1487-1502. doi:10.1128/MCB.00971-12.
- 25. PLoS ONE. 2013. 8 (6): e64953. doi:10.1371/journal.pone.0064953.
- 26. Nature Methods. 2013. 10 (7): 634-637. doi:10.1038/NMETH.2518.
- 27. Diabetes. 2013. 62(10): 3404–3417. doi:10.2337/db12-1650.
- 28. PLoS ONE. 2013. 8(7): e67513. doi:10.1371/journal.pone.0067513.
- 29. J. Clin. Invest. 2014. 124 (2):768-784. doi:10.1172/JCI69413.
- 30. PLoS ONE. 2014. 9(2): e89283. doi:10.1371/journal.pone.0089283.
- 31. Mol. Syst. Biol. 2014. 10(11): 762. doi:10.15252/msb.20145227.
- 32. PLoS ONE. 2014. 9(3): e91039. doi:10.1371/journal.pone.0091039.
- 33. Mol. Biosyst., 2015. 11 (3): 908-922 doi:10.1039/c4mb00490f.
- 34. Methods in Molecular Biology, 2015. 1295: 275-292. doi:10.1007/978-1-4939-2550-6 21.
- 35. PloS one. 2015. 10 (10): e0140619. doi:10.1371/journal.pone.0140619.
- 36. J. Proteomics. 2015. 128: 352-364. doi:10.1016/j.jprot.2015.08.015.
- 37. Plant Mitochondira: Methods and Protocols. 2015.107-121. doi:10.1007/978-1-4939-2639-8_7.
- 38. J Virol. 2016 Feb 3. pii: JVI.03175-15. doi:10.1128/JVI.03175-15.
- 39. PLoS ONE. 2015. 10(5): e0126242. doi:10.1371/journal.pone.0126242.
- 40. Nat Commun. 2015. 6: 7726. doi:10.1038/ncomms8726.
- 41. Nat Biotechnol. 2015. 33(4): 415-423. doi:10.1038/nbt.3130.
- 42. EMBO Rep. 2016. 17(3): 455-469. doi:10.15252/embr.201541132.
- 43. Arch. Biochem. Biophys. 2016. 598: 1-10. doi:10.1016/j.abb.2016.03.025.
- 44. Cancers. 2016. 8(3): 37. doi:10.3390/cancers8030037.
- 45. mSystems. 2016. 1(3): e00005-16. doi:10.1128/mSystems.00005-16.
- 46. J Biol Chem. 2016. 291(11): 5484-5499. doi:10.1074/jbc.M115.707091.
- 47. Sci Rep. 2016. 6: 19722. doi:10.1038/srep19722.
- 48. JCI Insight. 2016. 2(1): e84897. doi:10.1172/jci.insight.84897.
- 49. bioRxiv. 2016. 057174. doi:http://dx.doi.org/10.1101/057174.
- 50. Archives of Biochemistry and Biophysics. 1-10. 2016. 598.
- 51. Molecular & Cellular Proteomics. 2016. doi: 10.1074/mcp.O116.065219
- 52. American Society for Microbiology. 2016. 1(3): 1-19. doi: 10.1128/mSystems.00005-16.
- 53. Cancers. 2016. 8(3): 1-13. doi: 10.3390/cancers8030037.
- 54. JCI Insight. 2016. 1(2): 1-14. doi: 10.1172/jci.insight.84897.
- 55. Universitat zu Koln. 2016. 1-169.
- 56. EMBO Press. 2016. 17(3): 455-469. doi: 10.15252/embr.201541132.
- 57. Nature Biotechnology. 2016. 34(11): 1198-1205. doi: 10.1038/nbt.3681.
- 58. Journal of Visualized Experiments. 2016. 108. doi: 10.3791/53563.
- 59. BioRxiv. 2016. doi: https://doi.org/10.1101/057174.
- 60. Journal of The American Society for Mass Spectrometry. 2016. 27 (11) 1758-1771. doi: 10.1007/s13361-016-1476-zg.
- 61. The Journal of Biological Chemistry. 2016. 291 (11) 5484-5499 doi: 10.1074/jbc.M115.707091.
- 62. Molecular & Cellular Proteomics. 2016. 15 (2): 493-505. doi: 10.1074/mcp.M115.049288.
- 63. PNAS. 2018. 115 (1): 210-215. doi:10.1073/pnas.1717519115
- 64. Methods Mol Biol. 2015;1305:107-21. doi: 1007/978-1-4939-2639-8_7.
- 65. Mitochondrion. 2014 Nov;19 Pt B:252-60. doi: 10.1016/j.mito.2014.03.004
- 66. Protein Acetylation. 2019. 1983: 57-77. doi:10.1007/978-1-4939-9434-2_5
- 67. Molecular Metabolism. 2019. 25: 35-49. doi: 10.1016/j.molmet.2019.04.008
- 68. The Plant Journal. 2019. 99(1): 176-194. doi:10.1111/tpj.14315
- 69. Molecular Cell. 2019. 74(6): 1250-1263. doi: 10.1016/j.molcel.2019.04.009
- 70. Connective Tissue Research. 2019. doi: 10.1080/03008207.2019.1648443
|

 The acetylated peptide profile from acetyl-lysine-specific affinity chromatography (matrix = ICP0388). Approximately 5 mg of trypsinated crude protein from TSA treated MMRU cells were loaded.
The acetylated peptide profile from acetyl-lysine-specific affinity chromatography (matrix = ICP0388). Approximately 5 mg of trypsinated crude protein from TSA treated MMRU cells were loaded.